Wir beschleunigen Forschung auf industriellen Daten
Von der Forschungsdienstleistung für anspruchsvolle KI-Projekte bis zur notwendigen GPU- und Cloud-Infrastruktur. Das SDIL als “Datenreinraum” und zertifizierter BDVA iSpace schließt die Lücke zwischen akademischer Forschung und Industrieproblemen durch einen datengetriebenen Innovationszyklus.
Innovation in Ihrem Unternehmen umsetzen?
Starten Sie Ihr KI-Projekt in einem Transfersprint.
Gefördert vom BMBF. Begleitet von der Spitzenforschung.
Bewerben Sie sich jetzt.

Smart Data
Innovation Services
Gefördert vom BMBF baut das SDIL momentan einen Katalog mit “Innovation Services” auf. Diese können Unternehmen und Behörden nutzen um KI-Innovationen schneller auf den Weg zu bringen. Dank Förderung sogar bis Ende 2024 kostenlos für insgesamt 15 innovative Mikroprojekte.








Die SDIL-Plattform
Das Smart Data Innovation Lab (SDIL) ist eine Austausch- und Betriebsplattform, um die Kooperation zwischen Wirtschaft, öffentlicher Hand und Forschung im Bereich KI, Big Data und Smart Data-Technologien zu beschleunigen.
Unternehmen und deutsche Spitzenforschungseinrichtungen haben sich im SDIL organisiert und einen Rahmen zwischen den beteiligten Partnern vereinbart. Das Konzept für SDIL wurde in der Arbeitsgruppe „Bildung und Forschung für die digitale Zukunft“ des Nationalen IT-Gipfels der Bundesregierung schon 2014 entwickelt.
Die SDIL-Plattform ist mittlerweile als “Datenreinraum” etabliert innerhalb dessen auf Hochleistungsrechnern und mit rechtlicher und technischer Rahmen beschleunigte Co-Innovationen vorangetrieben werden können.
Hierzu betreibt das Steinbuch Centre for Computing (SCC) am KIT beispielsweise einen GPU-Cluster und bietet modernste Software und Hardware. Abgerundet wird das ganze durch aktuelle Cloud-Angebote der beteiligten Softwarepartner, die Niederschwellig im vorwettbewerblichen Bereich genutzt werden können.
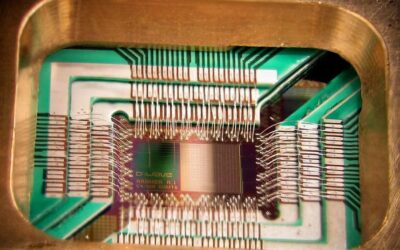
GPU Cluster
KI Cluster am KIT für Forschung auf Industriellen Daten mit 76 A100 GPU und Infiniband 100 Gbit Vernetzung
Datenreinraum
Sichere, zeitlich begrenzte Verarbeitung der Daten nach Bedarf on-premise am KIT oder bei Bedarf auch in der Cloud
Jupyter Lab
Interaktive sichere Python-Notebook-Umgebung mit vorinstallierter Software auf den Rechencluster
Vernetzung
Vernetzung und Einbindung der KI über Cloudressourcen
News & Events

WEB-SEMINAR: „KI und Nachhaltigkeit – AI and Sustainability“
In der heutigen Geschäftswelt sind Künstliche Intelligenz (KI) und Nachhaltigkeit mehr als bloße Schlagworte – sie sind entscheidende Faktoren für den langfristigen Erfolg Ihres Unternehmens. Möchten Sie erfahren, wie innovative KI-Technologien Ihre...
Smart Data Innovation Services: Edge and IoT Enablement for AI Applications
Datum: 14. Dezember 2023, 13:00 bis 16:00 Uhr Ort: online Diese Themen erwarten Sie: Vorstellung der Dienste von SAP und Software AG èFokus auf das Ausrollen von KI-Lösungen in der Anwendung Matchmaking und Diskussion mit KI-Anwendern: Produktion, Umwelt, Gesundheit …...
Unsere Referenzen
Von der Masterarbeit im Unternehmen, über den schnellen Forschungstranfer bis hin zum mehrjährigen kollaborativen Forschungsprojekt unterstützen wir KI-Forschung auf industriellen Datenquellen.
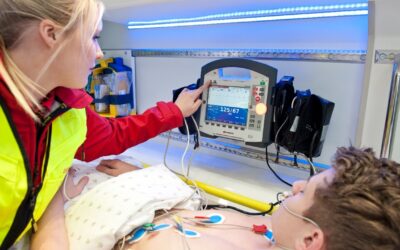
Optimierte Vorhersage des Defibrillationserfolgs mittels Few-Shot Learning
Mittels Few-Shot Learning soll eine präzise Vorhersage des Defibrillationserfolgs erreicht werden, um aussichtslose und damit potenziell schädliche Defibrillationen zu vermeiden.
SmartAPMotion
Die Fusion von maschinellen Lernverfahren verschiedene Sensor-Ströme und Datenquellen (unter anderem auch Video und Sprache) wird zur multimodale Aktivitätserkennung genutzt.
Conversational AI for Digital Twin
Nahtlos mit den Daten Ihres Unternehmens in natürlicher Sprache zu kommunizieren und datengesteuerte Entscheidungen zu treffen ermöglicht Ihnen der hier entwickelte KI-Assistent.
MangoTune: Adaption von CNN Modellen zur Reifevorhersage auf multi-spektralen Satellitendaten
Das Projekt bestimmt mittels KI-Analyse von multispektralen Satellitenbildern den Zeitbereich des Pflanzenwachstumsprozesses von Mangos weltweit und verknüpft das Ergebnis mit bestehenden Daten.
MAIWY – Erkennung von Weinblattkrankheiten mit KI
MAIWY strebt an, mithilfe von Smartphone-Bildern und künstlicher Intelligenz (KI) Rebkrankheiten, Mangelerscheinungen und Schädlinge bereits in frühen Stadien zu erkennen und zu differenzieren.
TruthfulLM: Verifying and Ensuring Truthfulness in Large Language Models
Die entwickelten Modelle extrahieren strukturierte Informationen aus Texten mit Hilfe derer generierte Texte automatisch mit einem Wissensgraphen abgeglichen und so auf ihren Wahrheitsgehalt überprüft werden können.
Feature Indexing für Parfumflaschen durch Deep Learning am Beispiel Parfümerie Pieper
Automatisierte Verbesserung der Parfümerie Pieper Produkt-Datenbank
WearNet: DeepLearning-Toolkit für Wearable-Bewegungsanalysen
Das Projekt befasst sich mit der Entwicklung modularer Deep Learning Systeme für Smart Clothing. Intelligente Kleidung, bei der die Elektronik in der textilen Struktur integriert ist, wird seit langem als zukunftsträchtig gesehen. Trotz des großen Interesses bleiben intelligente Textilien derzeit allerdings ein Nischenprodukt.
Chatbot Mining: Automatisierte Optimierung von Kundenservice-Chatbots
Kleine Energieversorger nutzen vermehrt Chatbots, doch mittelständische Unternehmen haben Probleme, die Chatbot-Daten zu analysieren, um ihren Kundenservice zu verbessern. Das Projekt zielt darauf ab, automatisierte Lösungen zu präsentieren, die die Potenziale dieser Daten erschließen.
Smart Air Quality Sensor Calibration
KI-gestützte Interpolation zur entfernten Kalibrierung von Feinstaub-Sensoren
TransPer: Transparente Personalisierung im E-Commerce
TransPer betrachtet Aspekte der Kausalität, Robustheit und Unsicherheit von KI-Anwendungen im industriellen Umfeld. Spezifischer Fokus: wie Produktempfehlungen im eCommerce-Bereich transparenter gestaltet werden können, um eine bessere Kundenzufriedenheit zu erreichen und die Einhaltung rechtlicher Vorgaben zu gewährleisten.
Verbesserung von KI Modellen basierend auf räumlichen Daten in der Agrarwirtschaft
Die Verbesserung der Bodenfeuchte-Vorhersagen für Heliopas durch eine effiziente Datenverarbeitungspipeline und Hyperparameteroptimierung
Nutzung maschinellen Lernens zur Zuordnung von Fragestellungen und Experten in Lernplattformen
Effektives Cognitive Matchmaking in Lernplattform-Communities: Untersuchung von LDA-Rec und SBERT-Rec Recommender-Systemen zur Experten-Fragen-Zuordnung
KI Modelle für Smart Cities am Beispiel Einzelhandel und Smart Parking in Dortmund
Die Erstellung eines flexiblen “Daten-Visualisierungs-Dashboards” und Vorhersagemodellen auf Basis von Autoregressionsmethoden
BERTI-4.0: Pre-trained Language Model für Conversational Agents im Kontext von Industrie 4.0
Intelligente Fabriken durch BERTI-4.0: Vortraining, Fine-Tuning und Evaluierung für Industrie 4.0 Sprachassistenzkomponenten
Nutzung von Quantum-Algorithmen für Konsistenzprüfung in Finanzberichten
Anwendbarkeit von Quantum-Computing auf Finanzberichte für effiziente Konsistenzprüfung
SDCS-BW: Price Intelligence
Die richtigen Algorithmen für erfolgreiche Multilabel-Produktklassifizierung aufspürt
Risk Management 4.0
Maschinellen Lernens zur vorausschauenden kausalen Analyse von Risikofaktoren über Investmentportfolien
„Cognitive Assistant“
Service-Assistent für Steuerberater, Wirtschaftsprüfer und Rechtsanwälte bei DATEV
SIS Software
Vorhersagen von Personenströmen, wenn nur kurzfristige Trainingsdaten zur Verfügung stehen
SDSC-BW: EDI
Die Position der Bemaßung detektieren und die Winkelinformationen der detektierten Positionen bereitzustellen
Verteilte Auswertung von hochdimensionaler Messdaten
Verteilte Auswertung hochfrequenter Messdaten aus der industriellen Fertigung für Qualitätsoptimierung und Condition Monitoring
SDCS-BW: ArtiMinds
Die Beziehung zwischen Startposition, Suchbahn des Roboterarms und durchschnittlichem Zeitaufwand
SDCS-BW: Schlötter
Intelligente Verkaufsmengen vorhersagen, um die durch ungenaue Prognosen verursachten Verluste zu verringern
Fraunhofer IOSB-INA
Evaluation von Verfahren zur automatischen Rekonfiguration von Transportdrohnen
E-Scooter Detection
Entwicklung neuer Technologien für die infrastrukturseitige Erfassung von Verkehrsdaten
SDSC-BW: Börse Stuttgart
Besseres Verständnis für komplexe Trading-Aktivitäten dank Transaktionsanalyse
SDCS-BW: Ensinger
Der ersten Schritt für eine bessere Verkaufsvorhersage, um mögliche Engpässe an heißen Tagen zu verhindern
QuestMiner
Erkennung und Bewertung von Anomalien in Grafiken
Churn-Warnsystem
Präzise und zeitnah Vorhersagen zu treffen, so dass das Unternehmen ausreichend Zeit hat, seine Kunden zu halten
SDSC-BW: Hectronic
Mit Smart-Data-Analysen verunreinigte Tankfüllstände frühzeitig erkennen
STEP
Ein Simulationsmodell für die Technikereinsatzplanung und Optimierung der Technikerrouten
SDCS-BW: BIA Forst
Eine Methode zur Ermittlung von Soll-Werten zur Verringerung der Schadenanzahl zeigen
SDCS-BW: BrandGroup
Eine genauere Einstellung der Messmerkmale und eine frühzeitige Identifizierung von Qualitätsabweichungen
SDSC-BW: LGI
Smarte Versandvolumenvorhersage mit KI-Modellen
SDCS-BW: DEHA Gruppe
Eine automatisierte Auswertung der Transaktionsdaten
BigGIS
Big Data und der Fusion unsicherer, geografischer Daten
SDSC-BW: Coral
Wissen besser vernetzen -Implementierung einer neuronalen Active-Learning-Netzwerkarchitektur
SDSC-BW: Vitra
Absatzprognosen von Form und Farbe – ein Vorhersagemodel auf Basis der Vertriebszahlen
billiger.de
Analyse von Nutzerverhalten basierend auf Web-Protokoll-Daten
SDSC-BW: Mader
Vorausschauend die Energieeffizienz steigern – eine energieeffiziente Optimierung des Kompressors
Transforming-Transport
Transforming Transport vereint Wissen und Lösungen großer europäischer ICT- und Big-Data-Technologieanbieter
Trelleborg Sealing Solutions
Zustandsüberwachung und -vorhersage von Dichtungssystemen
SmartAQnet
Entwicklung eines intelligenten, reproduzierbaren Messnetzwerkes in der Modellregion Augsburg
SDSC-BW: Erdrich
Dynamische Maschinenplanung mit Smart-Data-Technologien
OSRAM
Aus der Datenauswertung können Prognosen und Regeln für die Produktion erstellt werden
Erweiterung der ITS
Verbesserung der Verkehrsflussvorhersage mit Umgebungsmodellen
All-Time Parts Prediction
Lieferkette aufzubauen mittels IBM SPSS Modeler, kombiniert mit IBM SPSS CADS, Python und R-Komponenten
SDSC-BW: Streit
Von Daten zur Holzqualität über entstehende Daten an der Sägelinie bis hin zu den Verkaufsdaten
SDSC-BW: da-cons
In den Bilddateien Pflanzen zuverlässig zu erkennen
John Deere
Reduzierung der Nacharbeit und die Vermeidung von Fehlern bei der Fertigung von Traktoren
KIT-Facility Management
Bedarfsanalyse für energetische Baumaßnahmen auf Basis historischer Infrastrukturdaten
SDSC-BW: Dr.Hartmann
Smart-Data-Analysen für industrielle Prozesswassersysteme – Genauigkeit von mindestens 95% erreicht
SDSC-BW: Sedus
Prognosen für die Produktionsmengen von spezifischen Ausstattungsvarianten
SDSC-BW: Echobot
Die richtigen Algorithmen bei Big-Data-Projekten – sowohl hardware- als auch softwareseitige Optimierungspotentiale
SAP – Datenalyse
Untersuchung verschiedener Big Data Plattformen hinsichtlich ihrer Performance bei forensischer Datenanalyse
MedTrend1
Intelligente Datenvorhersage von Trends in der Medizin.
SDSC-BW: FUCHS
Auf der Suche nach unbekannten Korrelationen, um Optimierungspotenziale aufzuspüren
SDSC-BW: Hermle AG
Smart Data reduziert Wartungsintervalle von Fräsmaschinen
VDAR
Verteilte Dezentrale Autonome Regelungssysteme für Dezentrale Energiemärkte
SAP
Assoziationsregeln für Hochdimensionale Stammdaten
TRUMPF
Analyse hochvarianter, zeitlich dünn besiedelter und unmarkierter Datensätze zur Durchführung zustandsbasierter Wartung
SmartFactoryKL
Prädiktive Instandhaltungsdatenanalyse anhand SmartFactoryKL-generierter Daten
SDI-X
Smart Data Innovation-Prozesse, -Werkzeuge und -Betriebskonzepte
SDSC-BW: Rolf Benz
Potentialanalyse Lederverschnittoptimierung – eine automatisierte Aufstellung und Überprüfung von Hypothesen
SDSC-BW: Huber
Smart-Data-gestützte Kampagnen-Analysen für das Marketing – zukünftigen Marketingaktionen
ABB
Assoziationsanalyse für datengetriebene Dienstleistungen auf Basis Industrieller Log-Dateien
Smart Brain Data Analyse
Ansatz zum maschinellen Lernen für eine Hintergrundsegmentierung des 3D-Bildvolumens eines Gehirngewebeblocks
SDSC-BW: Herrenknecht
Smart-Data-Algorithmen unterstützen, bei der Planung die Liefergenauigkeit
SDSC-BW: Bilcare
Smarte Sensordaten unterstützen die Fertigung von Verpackungslösungen
KIT
GPU + In-Memory-Datenmanagement für Big Data-Analysen
Smart Data-Technologien
Smart Data-Lösungen für den produzierenden Mittelstand in Baden-Württemberg